P8: Graph Neural Network Based Multi-Agent Learning for Hierarchical Adaptive Coarse-Graining
Coarse-graining of molecular dynamics simulations is a powerful technique to bridge time and length scales that are often infeasible to study on an atomistic level. Next to classical coarse-graining simulations, machine learning models have been successfully used to predict the conditional free energy landscape with respect to the coarse-grained coordinates. However, most approaches are limited in their expressiveness due to linear mappings, utilize only a single coarse-graining resolution, and use a fixed training dataset of molecular dynamics snapshots.
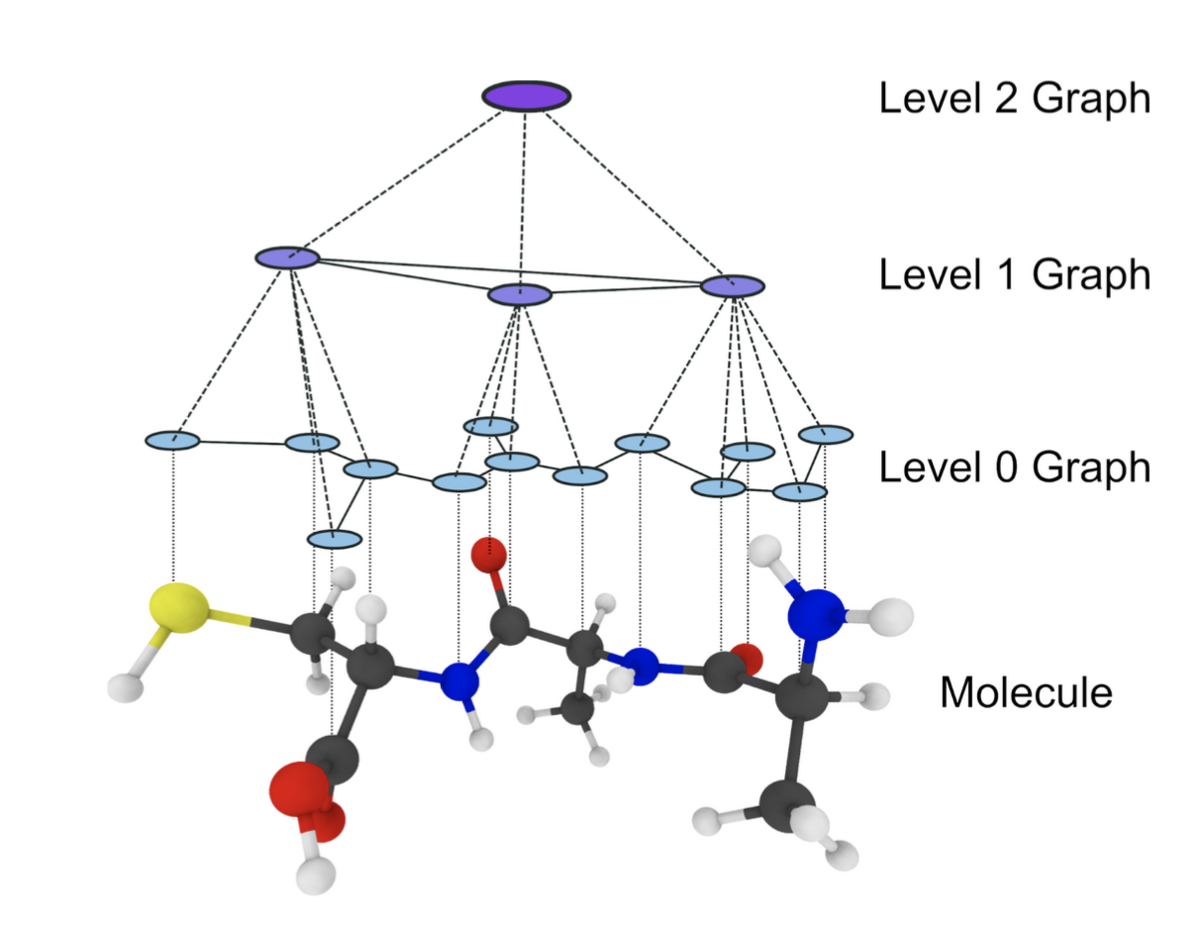
In this project, we develop hierarchical graph neural networks with invertible connections between resolution levels to study the dynamics of molecules on multiple scales simultaneously. Instead of the commonly used linear mapping operators, we use learned non-linear mappings in the form of invertible neural networks. This allows non-trivial internal collective degrees of freedom to be embedded in the latent space of the higher resolution levels. The invertible connections will allow probabilistic backmapping to the atomistic resolution, making the analysis on the atomistic level possible. This further allows us to use active learning methodology to gather more training data in high-error regions and to explore the configuration space in an efficient manner. Furthermore, higher levels of coarse-graining resolution allow the study of the collective motion of the molecule, making it possible to identify key collective variables.
| Name | Title | Group | |
|---|---|---|---|
| Schopmans, Henrik | Prof. Pascal Friederich (INT, KIT) | henrik schopmans ∂does-not-exist.kit edu |