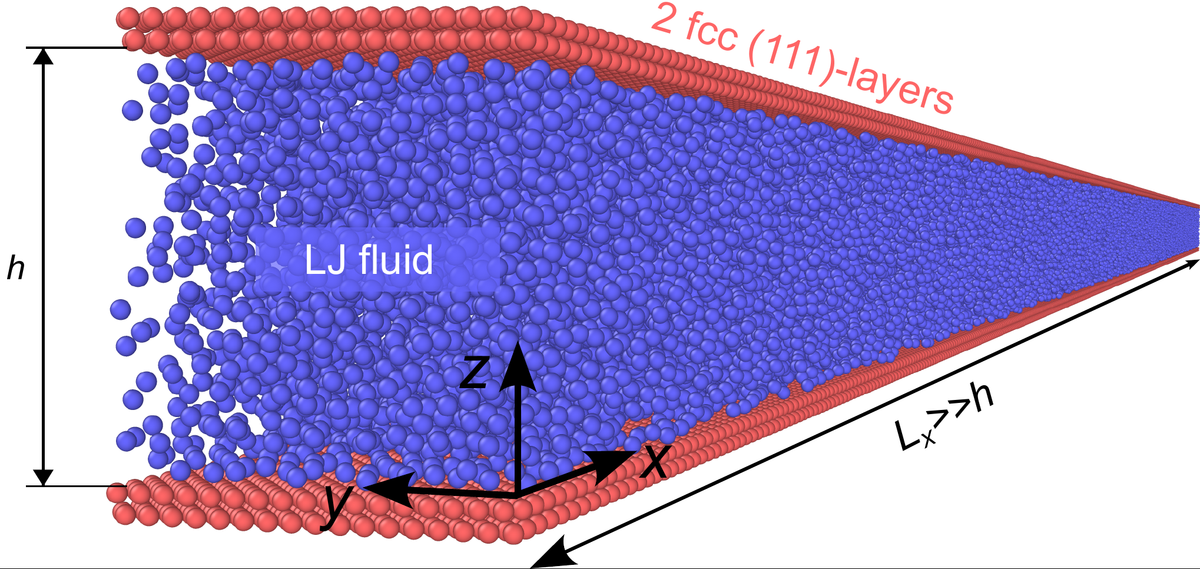
Hannes Holey, Peter Gumbsch, and Lars Pastewka.
"Confinement-Induced Diffusive Sound Transport in Nanoscale Fluidic Channels."
Phys. Rev. Lett. 131 (8), 2023. https://doi.org/10.1103/PhysRevLett.131.084001.
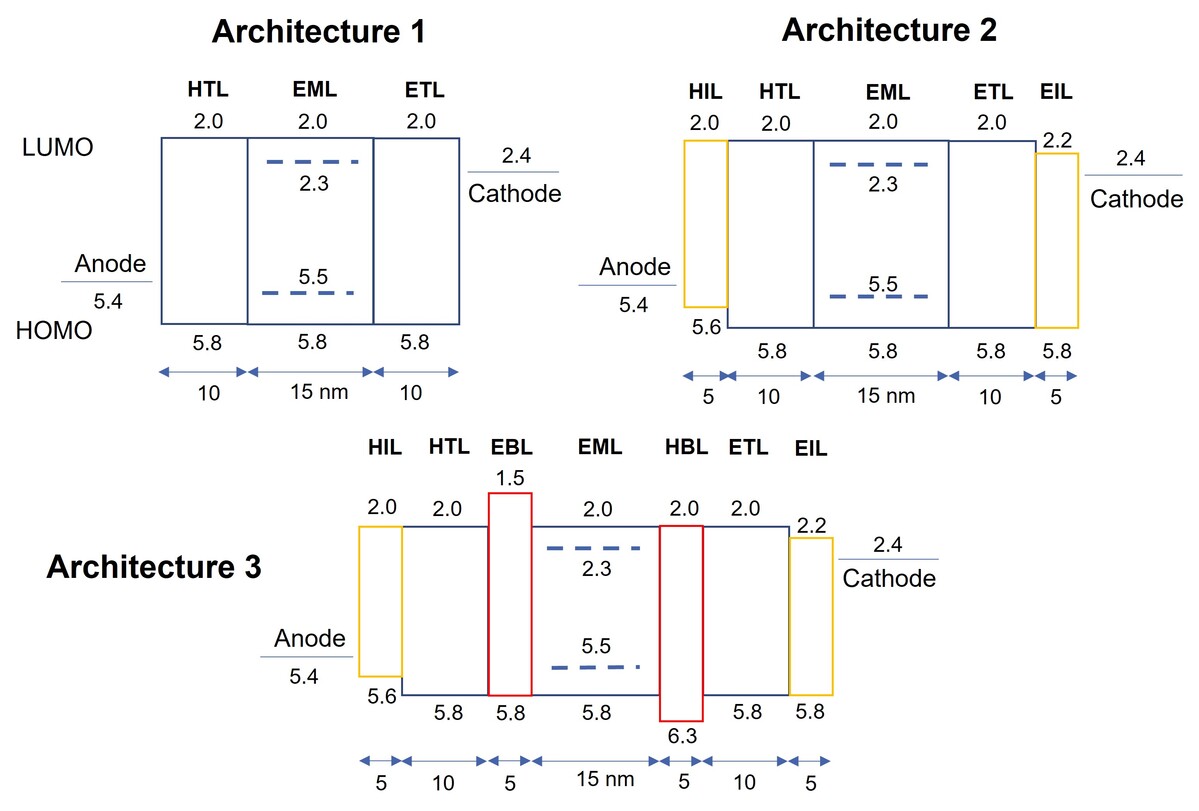
Özdemir, Ali Deniz, Fabian Li, Franz Symalla, and Wolfgang Wenzel.
“In Silico Studies of OLED Device Architectures Regarding Their Efficiency.”
Front. Phys. 11, 2023. https://www.frontiersin.org/articles/10.3389/fphy.2023.1222589.
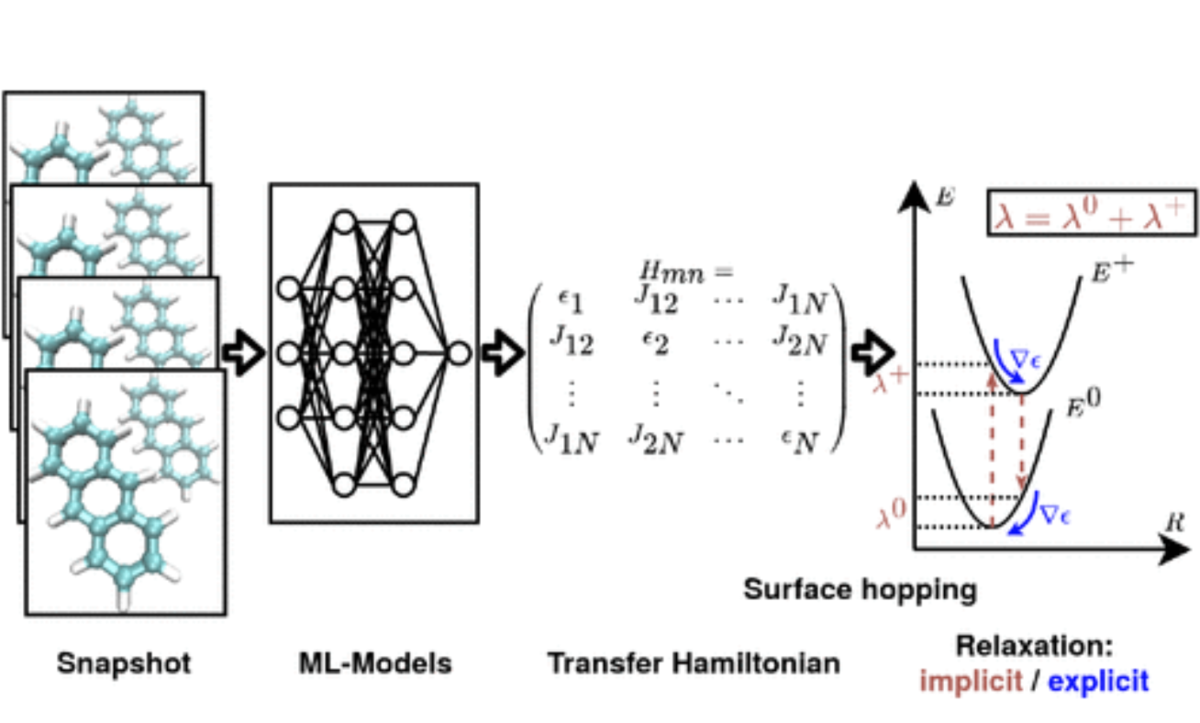
Dohmen, Philipp M., Mila Krämer, Patrick Reiser, Pascal Friederich, Marcus Elstner, and Weiwei Xie.
Modeling Charge Transport in Organic Semiconductors Using Neural Network Based Hamiltonians and Forces.
J. Chem. Theory Comput. 19, 13, 2023: 3825–38. https://doi.org/10.1021/acs.jctc.3c00264.
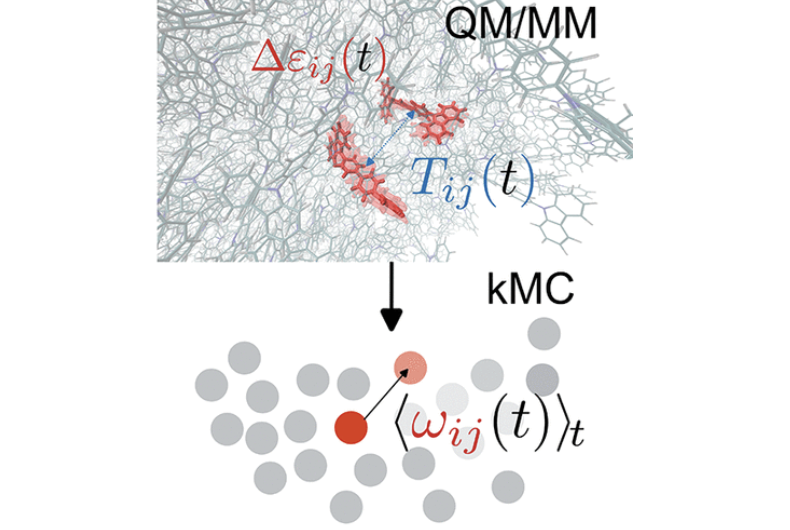
Özdemir, Deniz Ali, Samaneh Inanlou, Franz Symalla, Weiwei Xie, Wolfgang Wenzel, and Marcus Elstner.
Dynamic Effects on Hole Transport in Amorphous Organic Semiconductors: A Combined QM/MM and kMC Study.
J. Chem. Theory Comput., 2023. https://doi.org/10.1021/acs.jctc.3c00385.
Monzel, Laurenz, Christof Holzer, and Wim Klopper.
Natural Virtual Orbitals for the GW Method in the Random-Phase Approximation and Beyond.
J. Chem. Phys. 158, 14, 2023: 144102. https://doi.org/10.1063/5.0144469.
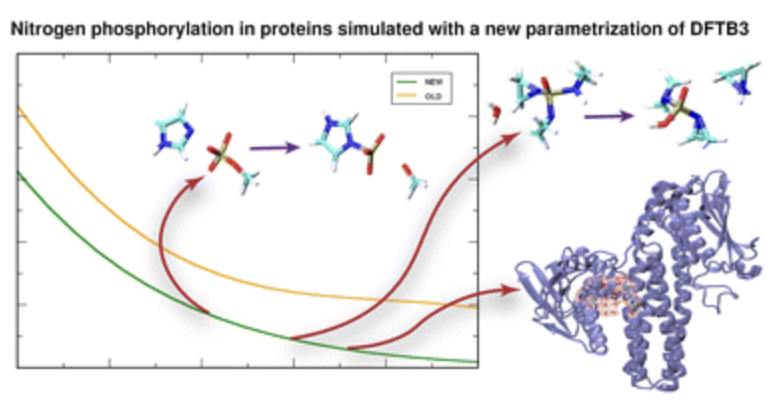
Kansari, Mayukh, Lena Eichinger, and Tomáš Kubař.
Extended-Sampling QM/MM Simulation of Biochemical Reactions Involving P–N Bonds.
Phys. Chem. Chem. Phys., 2023, 10.1039.D2CP05890A. https://doi.org/10.1039/D2CP05890A.
Schopmans, Henrik, Patrick Reiser, and Pascal Friederich.
Neural Networks Trained on Synthetically Generated Crystals Can Extract Structural Information from ICSD Powder X-Ray Diffractograms.
arXiv Preprint, 2023. https://arxiv.org/abs/2303.11699
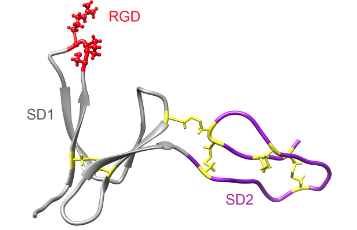
Kutzki, Fabian, Diego Butera, Angelina J. Lay, Denis Maag, Joyce Chiu, Heng-Giap Woon, Tomáš Kubař, Marcus Elstner, Camilo Aponte-Santamaría, Philip J. Hogg and Frauke Gräter.
Disulfide Bond Reduction and Exchange in Von-Willebrand-Factor’s C4-Domain Undermines Platelet Binding.
Journal of Thrombosis and Haemostasis, 2023. https://doi.org/10.1016/j.jtha.2023.03.039.
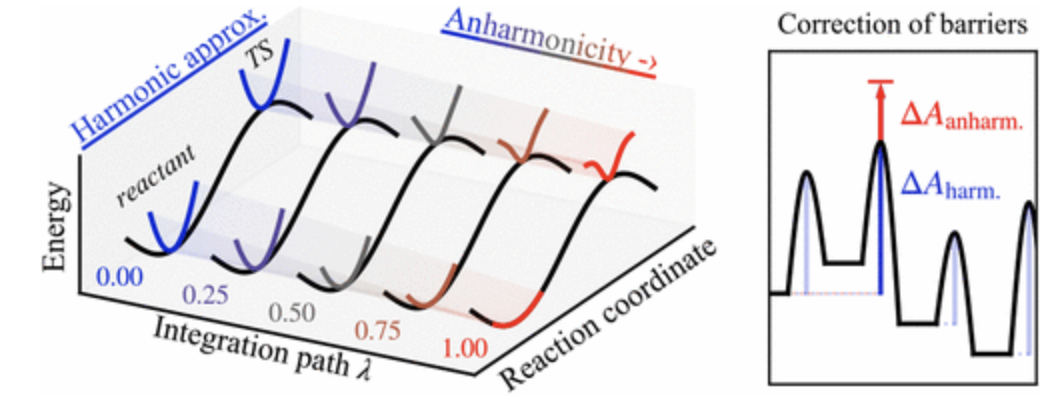
Amsler, Jonas, Philipp N. Plessow, Felix Studt, and Tomáš Bučko.
Anharmonic Correction to Free Energy Barriers from DFT-Based Molecular Dynamics Using Constrained Thermodynamic Integration
J. Chem. Theory Comput., 2023. https://doi.org/10.1021/acs.jctc.3c00169.
Esmaeilpour, Meysam, Saibal Jana, Hongjiao Li, Mohammad Soleymanibrojeni, and Wolfgang Wenzel.
A Solution-Mediated Pathway for the Growth of the Solid Electrolyte Interphase in Lithium-Ion Batteries.
Adv. Energy Mater. 13, no. 14 (2023): 2203966. https://doi.org/10.1002/aenm.202203966.
Schweer, Julie and Marcus Elstner.
Dealing with Molecular Complexity. Atomistic Computer Simulations and Scientific Explanation
Perspect. Sci., 2023, 1–37. https://doi.org/10.1162/posc_a_00594.
Maag, Denis, Josua Böser, Henryk A. Witek, Ben Hourahine, Marcus Elstner, and Tomáš Kubař.
Mechanism of Proton-Coupled Electron Transfer Described with QM/MM Implementation of Coupled-Perturbed Density-Functional Tight-Binding.
J. Chem. Phys. 158, 2023: 124107. https://doi.org/10.1063/5.0137122.
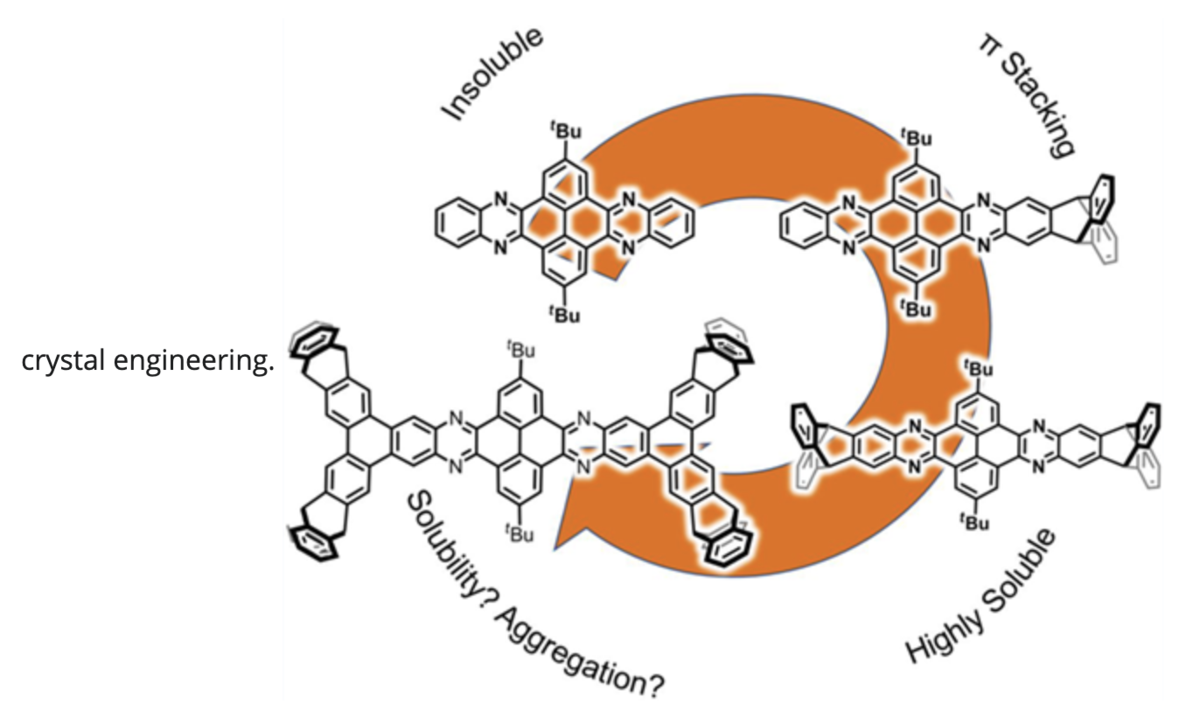
Roß, Lisa, Julius Reitemeier, Farhad Ghalami, Wen-Shan Zhang, Jürgen H. Gross, Frank Rominger, Sven M. Elbert, Rasmus Schröder, Marcus Elstner, and Michael Mastalerz.
Two Dimensional Triptycene End-Capping and Its Influence on the Self-Assembly of Quinoxalinophenanthrophenazines.
Chin. J. Chem., 41, 2023: 1198—1208. https://doi.org/10.1002/cjoc.202200754.
Grimm, Volker, Tobias Kliesch, and G. R. W. Quispel.
Discrete Gradients in Short-Range Molecular Dynamics Simulations.
arXiv preprint, 2023. https://arxiv.org/pdf/2212.14344.pdf
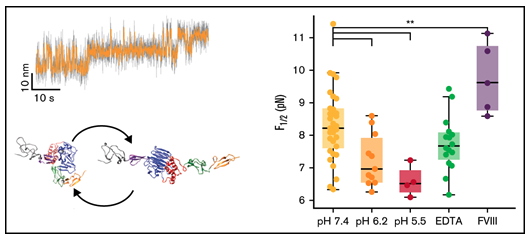
Gruber, Sophia, Achim Löf, Adina Hausch, Fabian Kutzki, Res Jöhr, Tobias Obser, Gesa König, et al.
A Conformational Transition of the D′D3 Domain Primes von Willebrand Factor for Multimerization.
Blood Adv 6 (17), 2022: 5198–5209. https://doi.org/10.1182/bloodadvances.2022006978.
Kutzki, Fabian, Diego Butera, Angelina J. Lay, Denis Maag, Joyce Chiu, Heng-Giap Woon, Tomáš Kubař, et al.
Dynamic Disulfide Bond Topologies in Von-Willebrand-Factor’s C4-Domain Undermine Platelet Binding.
bioRxiv preprint, 08, 2022. https://doi.org/10.1101/2022.08.20.504523
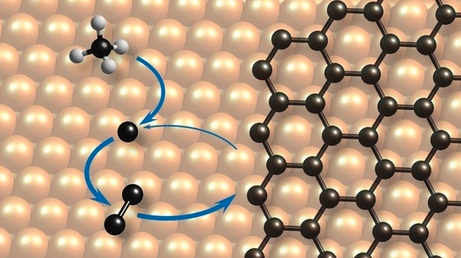
Popov, I., Bügel. P., Kozlowska, M., Fink, K., Studt, F., Sharapa, D. I.:
Analytical Model of CVD Growth of Graphene on Cu (111) Surface,
Nanomaterials 2022, 12 (17), 2963. https://doi.org/10.3390/nano12172963
Roozmeh, M.; Kondov, I.
Automating and Scaling Task-Level Parallelism of Tightly Coupled Models via Code Generation.
Computational Science – ICCS 2022. Groen, D., de Mulatier, C., Paszynski, M., Krzhizhanovskaya, V. V., Dongarra, J. J., Sloot, P. M. A., Eds.; Lecture Notes in Computer Science; Springer International Publishing: Cham, 2022; Vol. 13353, pp 69–82. https://doi.org/10.1007/978-3-031-08760-8_6.
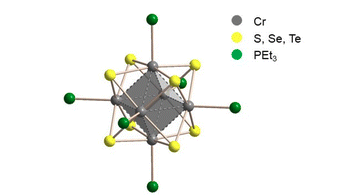
Bügel, P., Krummenacher, I., Weigend, F., Eichhöfer, A.:
Experimental and theoretical evidence for low-lying excited states in [Cr6E8(PEt3)6] (E = S, Se, Te) cluster molecules
Dalton Trans. 2022, 51, 14568–14580. https://doi.org/10.1039/D2DT01690G
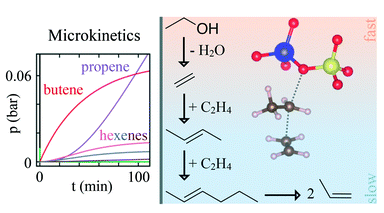
Amsler, J.,Bernart, S., Plessow, P. N., Studt, F.:
Theoretical investigation of the olefin cycle in H-SSZ-13 for the ethanol-to-olefins process using ab initio calculations and kinetic modeling
Catal. Sci. Technol., 2022, 12, 3311-3321. https://doi.org/10.1039/D1CY02289J
Schneider, S.; Bytyqi, K.; Kohaut, S.; Bügel, P.; Weinschenk, B.; Marz, M.; Kimouche, A.; Fink, K.; Hoffmann-Vogel, R.
Molecular self-assembly of DBBA on Au(111) at room temperature.
Phys. Chem. Chem. Phys. 2022, 28371–28380. doi:10.1039/D2CP02268K
Böser, Julian; Kubař, Tomáš; Elstner, Marcus; Maag, Denis.
Reduction Pathway of Glutaredoxin 1 Investigated with QM/MM Molecular Dynamics Using a Neural Network Correction.
J. Chem. Phys. 2022, 157(15): 154104. https://doi.org/10.1063/5.0123089.
Mostaghimi, Mersad; Rêgo, Celso R. C.; Haldar, Ritesh; Wöll, Christof; Wenzel, Wolfgang; Kozlowska, Mariana.
Automated Virtual Design of Organic Semiconductors Based on Metal-Organic Frameworks.
Front. Mater. 2022, 9. https://doi.org/10.3389/fmats.2022.840644
Suyetin, Mikhail; Bag, Saientan; Anand, Priya; Borkowska-Panek, Monika; Gußmann, Florian; Brieg, Martin; Fink, Karin; Wenzel, Wolfgang.
Modelling Peptide Adsorption Energies on Gold Surfaces with an Effective Implicit Solvent and Surface Model.
J. Colloid Interface Sci. 2022, 605, 493–99. https://doi.org/10.1016/j.jcis.2021.07.090.
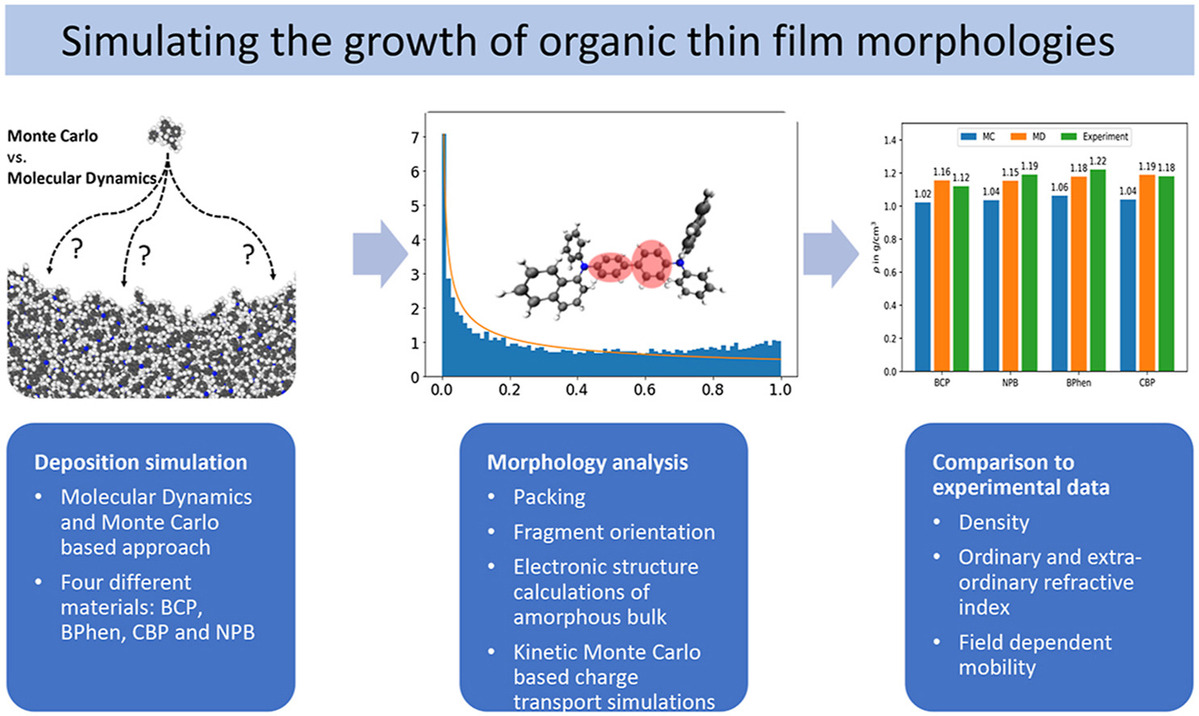
Degitz, C.; Konrad, M.; Kaiser, S.; Wenzel, W.
Simulating the Growth of Amorphous Organic Thin Films.
Org. Electron. 2022, 102, 106439. https://doi.org/10.1016/j.orgel.2022.106439.
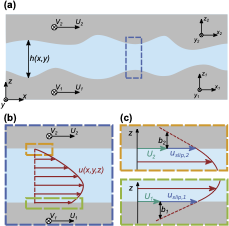
Holey, H.; Codrignani, A.; Gumbsch, P.; Pastewka, L.
Height-Averaged Navier–Stokes Solver for Hydrodynamic Lubrication.
Tribol. Lett. 2022, 70 (2), 36. https://doi.org/10.1007/s11249-022-01576-5.
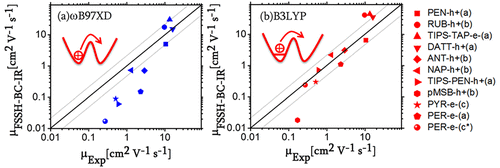
Roosta, S.; Galami, F.; Elstner, M.; Xie, W.
Efficient Surface Hopping Approach for Modeling Charge Transport in Organic Semiconductors.
J. Chem. Theory Comput. 2022, acs.jctc.1c00944. https://doi.org/10.1021/acs.jctc.1c00944.
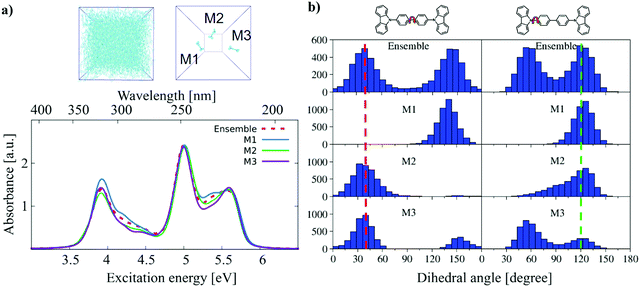
Inanlou, S.; Cortés-Mejía, R.; Özdemir, A. D.; Höfener, S.; Klopper, W.; Wenzel, W.; Xie, W.; Elstner, M.
Understanding Excited State Properties of Host Materials in OLEDs: Simulation of Absorption Spectrum of Amorphous 4,4-Bis(Carbazol-9-Yl)-2,2-Biphenyl (CBP).
Phys. Chem. Chem. Phys. 2022, 10.1039.D1CP04293A. https://doi.org/10.1039/D1CP04293A.
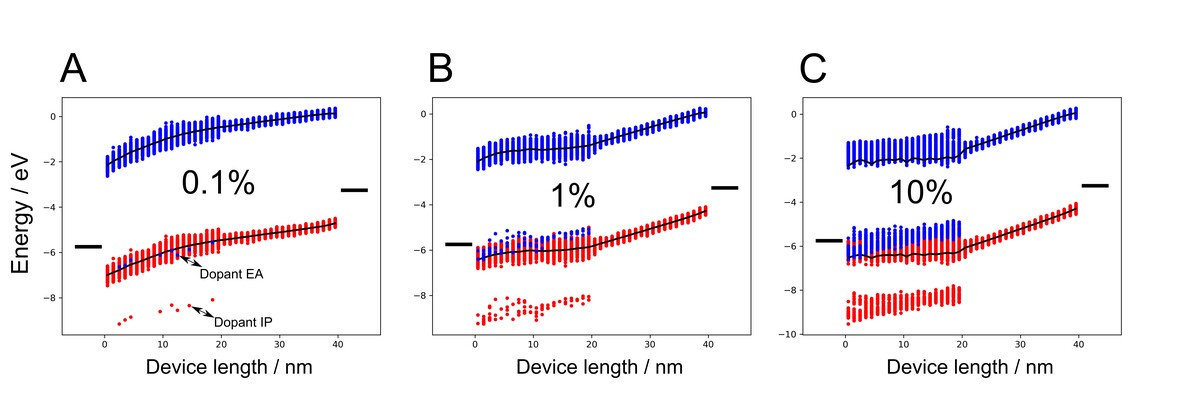
Özdemir, A. D.; Kaiser, S.; Neumann, T.; Symalla, F.; Wenzel, W.
Systematic kMC Study of Doped Hole Injection Layers in Organic Electronics.
Front. Chem. 2022, 9. https://doi.org/10.3389/fchem.2021.809415.
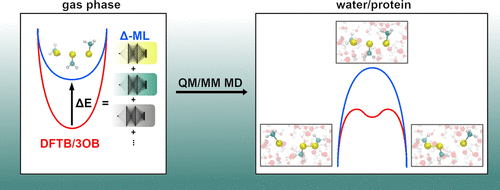
Gómez-Flores, C. L.; Maag, D.; Kansari, M.; Vuong, V.-Q.; Irle, S.; Gräter, F.; Kubař, T.; Elstner, M.
Accurate Free Energies for Complex Condensed-Phase Reactions Using an Artificial Neural Network Corrected DFTB/MM Methodology.
J. Chem. Theory Comput. 2022, acs.jctc.1c00811. https://doi.org/10.1021/acs.jctc.1c00811.
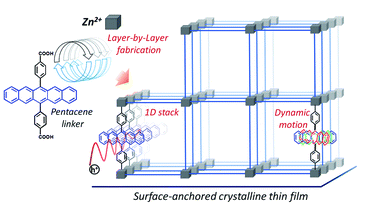
Haldar, Ritesh, Mariana Kozlowska, Michael Ganschow, Samrat Ghosh, Marius Jakoby, Hongye Chen, Farhad Ghalami, et al.
Interplay of Structural Dynamics and Electronic Effects in an Engineered Assembly of Pentacene in a Metal–Organic Framework.
Chem. Sci. 12 (12), 2021: 4477–83. https://doi.org/10.1039/D0SC07073D
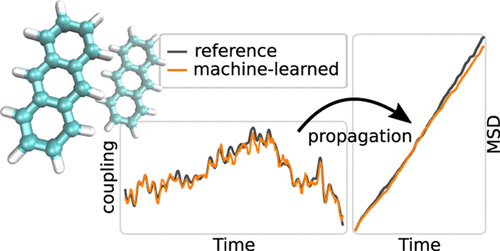
Schieschke, Nils, Beatrix M. Bold, Philipp M. Dohmen, Daniel Wehl, Marvin Hoffmann, Andreas Dreuw, Marcus Elstner, and Sebastian Höfener.
Geometry Dependence of Excitonic Couplings and the Consequences for Configuration-Space Sampling.
J Comput Chem. 42 (20), 2021: 1402–18. https://doi.org/10.1002/jcc.26552
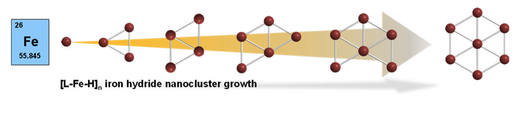
Chakraborty, U.; Bügel, P.; Fritsch, L.; Weigend, F.; Bauer, M.; Jacobi von Wangelin, A.
Planar Iron Hydride Nanoclusters: Combined Spectroscopic and Theoretical Insights into Structures and Building Principles.
Chemistry Open 2021, 10, 265–271. DOI doi:10.1002/open.202000307
Huck, Volker, Po-Chia Chen, Emma-Ruoqi Xu, Alexander Tischer, Ulrike Klemm, Camilo Aponte-Santamaría, Christian Mess, et al.
Gain-of-Function Variant p.Pro2555Arg of von Willebrand Factor Increases Aggregate Size through Altering Stem Dynamics.
Thromb Haemost, 122(02), 2021, 226–39. https://doi.org/10.1055/a-1344-4405.
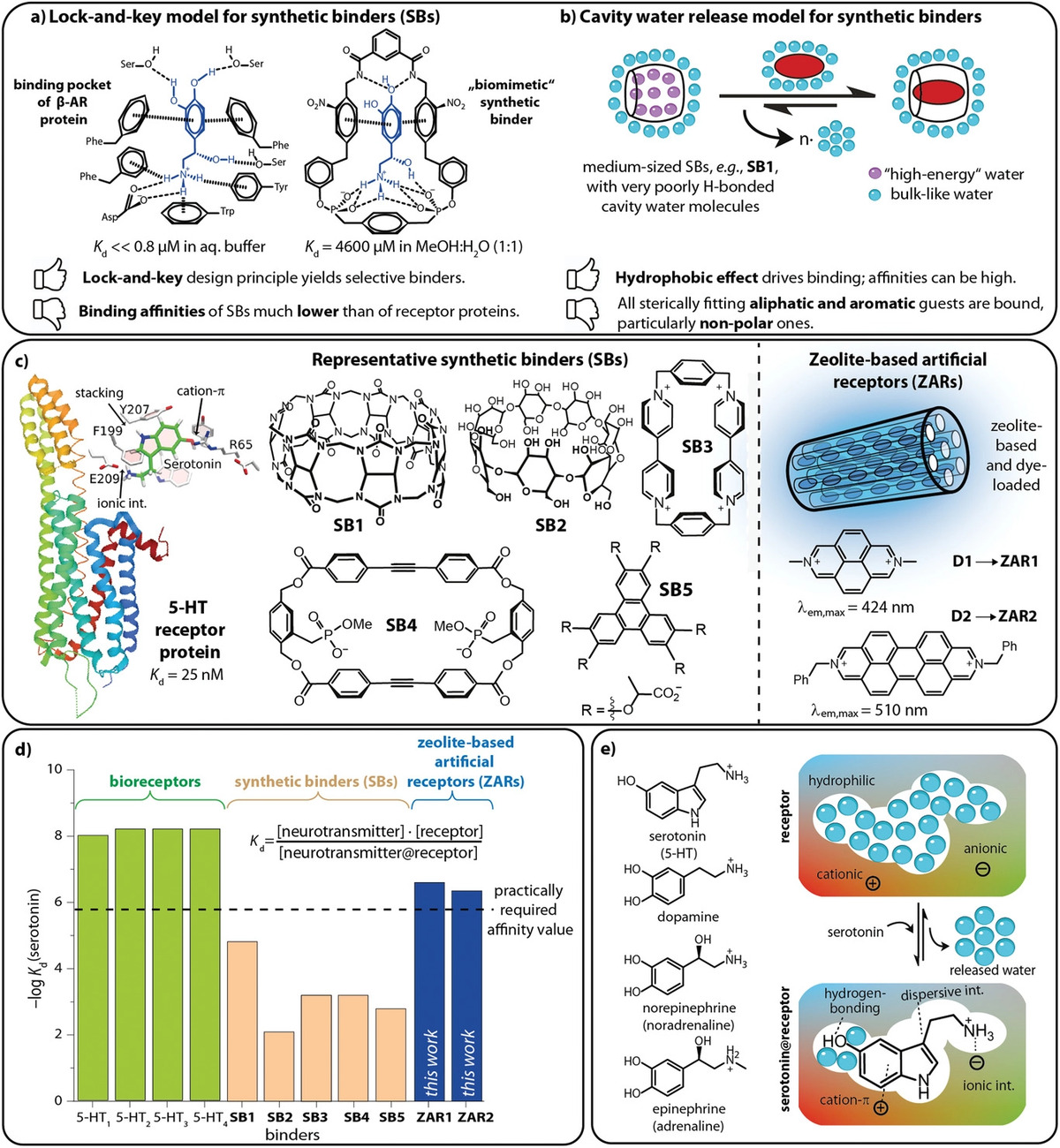
Grimm, Laura M.; Sinn, Stephan; Krstić, Marjan; D’Este, Elisa; Sonntag, Ivo; Prasetyanto, Eko Adi; Kuner, Thomas; Wenzel, Wolfgang; De Cola, Luisa; Biedermann, Frank.
Fluorescent Nanozeolite Receptors for the Highly Selective and Sensitive Detection of Neurotransmitters in Water and Biofluids.
Advanced Materials 2021, 33, 49: 2104614. https://doi.org/10.1002/adma.202104614.
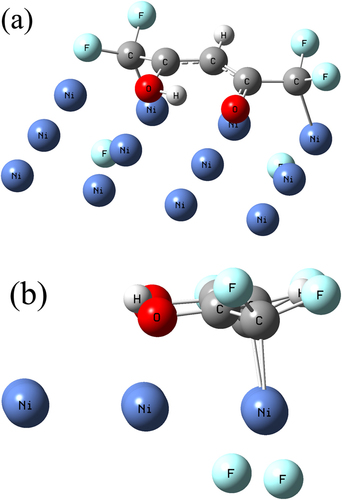
Basher, Abdulrahman H.; Krstić, Marjan; Fink, Karin; Ito, Tomoko; Karahashi, Kazuhiro;
Wenzel, Wolfgang; Hamaguchi, Satoshi.
Erratum: ‘Formation and Desorption of Nickel Hexafluoroacetylacetonate
Ni(Hfac)2 on a Nickel Oxide Surface in Atomic Layer Etching Processes’ [J.
Vac. Sci. Technol. A 38, 052602 (2020)].
J. Vac. Sci. Technol. A 2021, 39, 5: 057001. https://doi.org/10.1116/6.0001319.
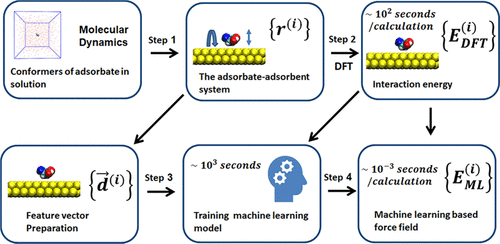
Bag, Saientan; Konrad, Manuel; Schlöder, Tobias; Friederich, Pascal; Wenzel, Wolfgang.
Fast Generation of Machine Learning-Based Force Fields for Adsorption Energies.
J. Chem. Theory Comput. 2021, 17, 11: 7195-7202. https://doi.org/10.1021/acs.jctc.1c00506.
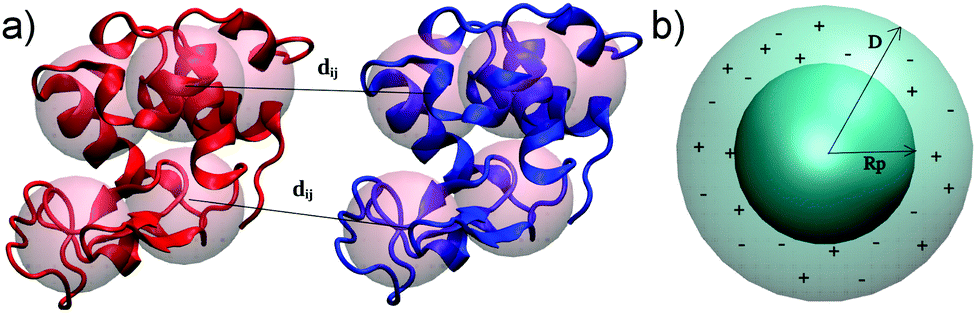
Pusara, Srdjan; Yamin, Peyman; Wenzel, Wolfgang; Krstić, Marjan; Kozlowska, Mariana.
A Coarse-Grained xDLVO Model for Colloidal Protein–Protein Interactions.
Phys. Chem. Chem. Phys. 2021, 23: 12780–94. https://doi.org/10.1039/D1CP01573G.
Özdemir, A. D.; Barua, P.; Pyatkov, F.; Hennrich, F.; Chen, Y.; Wenzel, W.; Krupke, R.; Fediai, A.
Contact Spacing Controls the On-Current for All-Carbon Field Effect Transistors.
Commun. Phys. 2021, 4 (1), 246. https://doi.org/10.1038/s42005-021-00747-5.
Maag, D.; Putzu, M.; Gómez-Flores, C. L.; Gräter, F.; Elstner, M.; Kubař, T.
Electrostatic Interactions Contribute to the Control of Intramolecular Thiol–Disulfide Isomerization in a Protein.
Phys. Chem. Chem. Phys. 2021, 10.1039.D1CP03129E. https://doi.org/10.1039/D1CP03129E.
Maag, D.; Mast, T.; Elstner, M.; Cui, Q.; Kubař, T.
O to BR Transition in Bacteriorhodopsin Occurs through a Proton Hole Mechanism.
Proc. Natl. Acad. Sci. USA 2021, 118 (39). https://doi.org/10.1073/pnas.2024803118.
Reiser, P.; Konrad, M.; Fediai, A.; Léon, S.; Wenzel, W.; Friederich, P.
Analyzing Dynamical Disorder for Charge Transport in Organic Semiconductors via Machine Learning.
J. Chem. Theory Comput. 2021, 17 (6), 3750–3759. https://doi.org/10.1021/acs.jctc.1c00191.
Konrad, M.; Wenzel, W.
CONI-Net: Machine Learning of Separable Intermolecular Force Fields.
J. Chem. Theory Comput. 2021, acs.jctc.1c00328. https://doi.org/10.1021/acs.jctc.1c00328.
Weiel, M.; Götz, M.; Klein, A.; Coquelin, D.; Floca, R.; Schug, A.
Dynamic particle swarm optimization of biomolecular simulation parameters with flexible objective functions.
Nat. Mach. Intell. 2021. https://doi.org/10.1038/s42256-021-00366-3.
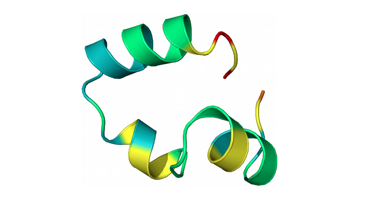
Christiansen, A.; Weiel, M.; Winkler, A.; Schug, A.; Reinstein, J.
The Trimeric Major Capsid Protein of Mavirus Is Stabilized by Its Interlocked N-Termini Enabling Core Flexibility for Capsid Assembly.
J. Mol. Biol. 2021, 166859. https://doi.org/10.1016/j.jmb.2021.166859.
Amsler, J.; Plessow, P. N.; Studt, F.; Bučko, T.
Anharmonic Correction to Adsorption Free Energy from DFT-Based MD Using Thermodynamic Integration.
J. Chem. Theory Comput. 2021,17 (2), 1155–1169. https://doi.org/10.1021/acs.jctc.0c01022.
Cortés-Mejía, R.; Höfener, S.; Klopper, W.
Effects of Rotational Conformation on Electronic Properties of 4,4′-Bis(Carbazol-9-Yl)Biphenyl (CBP): The Single-Molecule Picture and Beyond.
Mol. Phys. 2021, e1876936. https://doi.org/10.1080/00268976.2021.1876936.
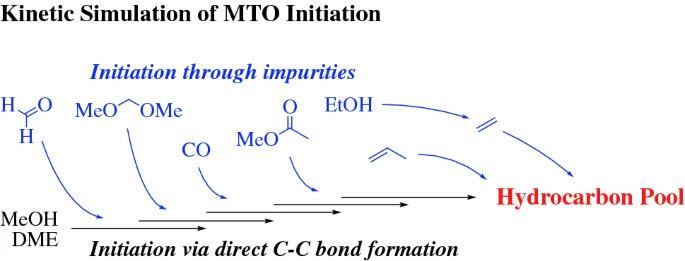
Amsler, J.; Plessow, P. N.; Studt, F.
Effect of Impurities on the Initiation of the Methanol-to-Olefins Process: Kinetic Modeling Based on Ab Initio Rate Constants.
Catal. Lett. 2021. https://doi.org/10.1007/s10562-020-03492-6.
Bold, Beatrix M., Monja Sokolov, Sayan Maity, Marius Wanko, Philipp M. Dohmen, Julian J. Kranz, Ulrich Kleinekathöfer, Sebastian Höfener, and Marcus Elstner.
Benchmark and Performance of Long-Range Corrected Time-Dependent Density Functional Tight Binding (LC-TD-DFTB) on Rhodopsins and Light-Harvesting Complexes.
Phys. Chem. Chem. Phys. 22,19 (2020): 10500–518. https://doi.org/10.1039/C9CP05753F
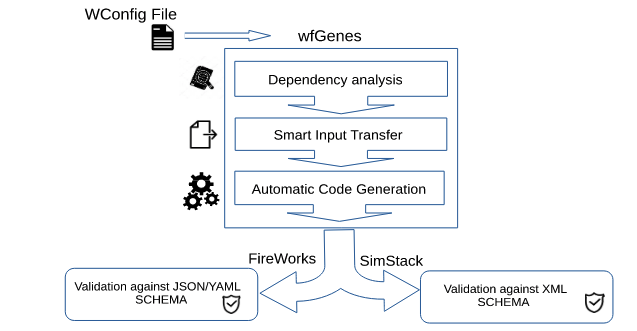
Roozmeh, M.; Kondov, I.
Workflow Generation with WfGenes.
IEEE/ACM workflows in support of large-scale science (WORKS). IEEE, 2020; pp 9–16. https://doi.org/10.1109/WORKS51914.2020.00007.
Amsler J., Sarma, B. B., Agostini, G., Prieto, G., Plessow, P. N., & Studt, F.:
Prospects of heterogeneous hydroformylation with supported single atom catalysts
J. Am. Chem. Soc., 2020,142 (11), 5087-5096. https://doi.org/10.1021/jacs.9b12171
Symalla, Franz; Heidrich, Shahriar; Friederich, Pascal; Strunk, Timo; Neumann, Tobias; Minami, Daiki; Jeong, Daun; Wenzel, Wolfgang.
Multiscale Simulation of Photoluminescence Quenching in Phosphorescent OLED Materials.
Adv. Theory Simul. 2020, 3, 4: 1900222. https://doi.org/10.1002/adts.201900222.
Basher, Abdulrahman H.; Krstić, Marjan; Takeuchi, Takae; Isobe, Michiro; Ito, Tomoko; Kiuchi, Masato; Karahashi, Kazuhiro; Wenzel, Wolfgang; Hamaguchi, Satoshi.
Stability of Hexafluoroacetylacetone Molecules on Metallic and Oxidized Nickel Surfaces in Atomic-Layer-Etching Processes.
J. Vac. Sci. Technol. A 2020, 38, 2: 022610. https://doi.org/10.1116/1.5127532.
Voronin, A.; Weiel, M.; Schug, A.
Including Residual Contact Information into Replica-Exchange MD Simulations Significantly Enriches Native-like Conformations.
PLoS ONE 2020, 15 (11), e0242072. https://doi.org/10.1371/journal.pone.0242072.
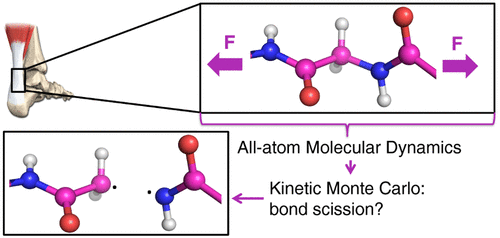
Rennekamp, B.; Kutzki, F.; Obarska-Kosinska, A.; Zapp, C.; Gräter, F.
Hybrid Kinetic Monte Carlo/Molecular Dynamics Simulations of Bond Scissions in Proteins.
J. Chem. Theory Comput. 2020, 16 (1), 553–563. https://doi.org/10.1021/acs.jctc.9b00786.
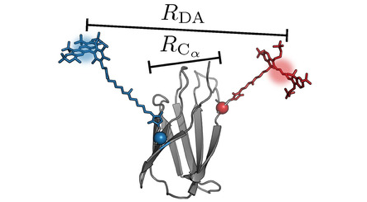
Reinartz, I.; Weiel, M.; Schug, A.
FRET Dyes Significantly Affect SAXS Intensities of Proteins.
Isr. J. Chem. 2020, 60 (7), 725–734. https://doi.org/10.1002/ijch.202000007.
Krämer, M.; Dohmen, P. M.; Xie, W.; Holub, D.; Christensen, A. S.; Elstner, M.
Charge and Exciton Transfer Simulations Using Machine-Learned Hamiltonians.
J. Chem. Theory Comput. 2020, 16 (7), 4061–4070. https://doi.org/10.1021/acs.jctc.0c00246.
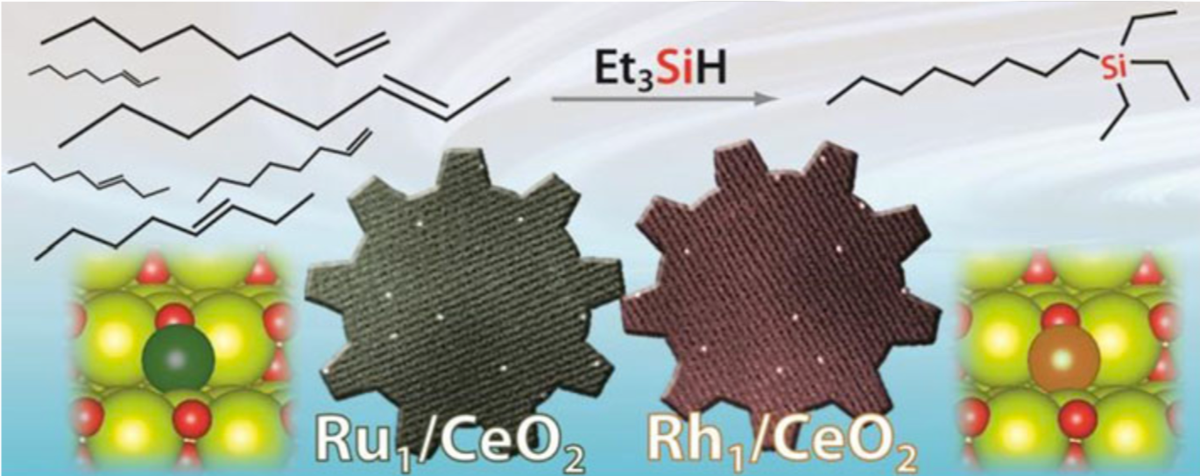
Sarma, B. B.; Kim, J.; Amsler, J.; Agostini, G.; Weidenthaler, C.; Pfänder, N.; Arenal, R.; Concepción, P.; Plessow, P.; Studt, F.; Prieto, G.
One‐Pot Cooperation of Single‐Atom Rh and Ru Solid Catalysts for a Selective Tandem Olefin Isomerization‐Hydrosilylation Process.
Angew. Chem. Int. Ed. 2020, 59 (14), 5806–5815. https://doi.org/10.1002/anie.201915255.
Amsler, J.; Sarma, B. B.; Agostini, G.; Prieto, G.; Plessow, P. N.; Studt, F.
Prospects of Heterogeneous Hydroformylation with Supported Single Atom Catalysts.
J. Am. Chem. Soc. 2020, 142 (11), 5087–5096. https://doi.org/10.1021/jacs.9b12171.
Weiel, M.; Reinartz, I.; Schug, A.
Rapid Interpretation of Small-Angle X-Ray Scattering Data.
PLoS Comput. Biol. 2019, 15 (3), e1006900. https://doi.org/10.1371/journal.pcbi.1006900.